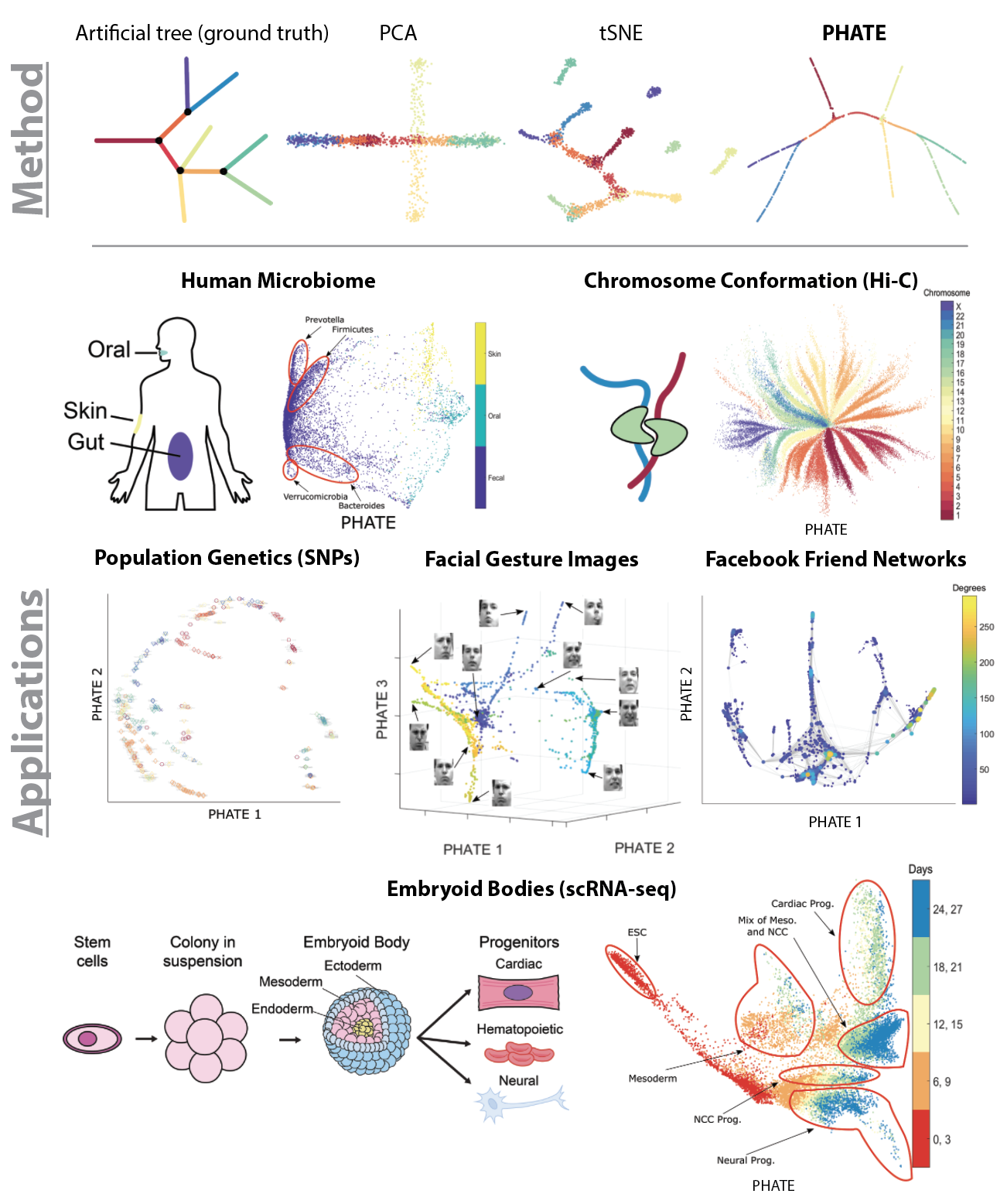
PHATE
Visualizing Transitions and Structure for Biological Data Exploration
In the era of 'Big Data' there is a pressing need for tools that provide human interpretable visualizations of emergent patterns in high-throughput high-dimensional data. Further, to enable insightful data exploration, such visualizations should faithfully capture and emphasize emergent structures and patterns without enforcing prior assumptions on the shape or form of the data.
In this paper, we present PHATE (Potential of Heat-diffusion for Affinity-based Transition Embedding) - an unsupervised low-dimensional embedding for visualization of data that is aimed at solving these issues. Unlike previous methods that are commonly used for visualization, such as PCA and tSNE, PHATE is able to capture and highlight both local and global structure in the data.