SAUCIE
Exploring Single-Cell Data with Deep Multitasking Neural Networks
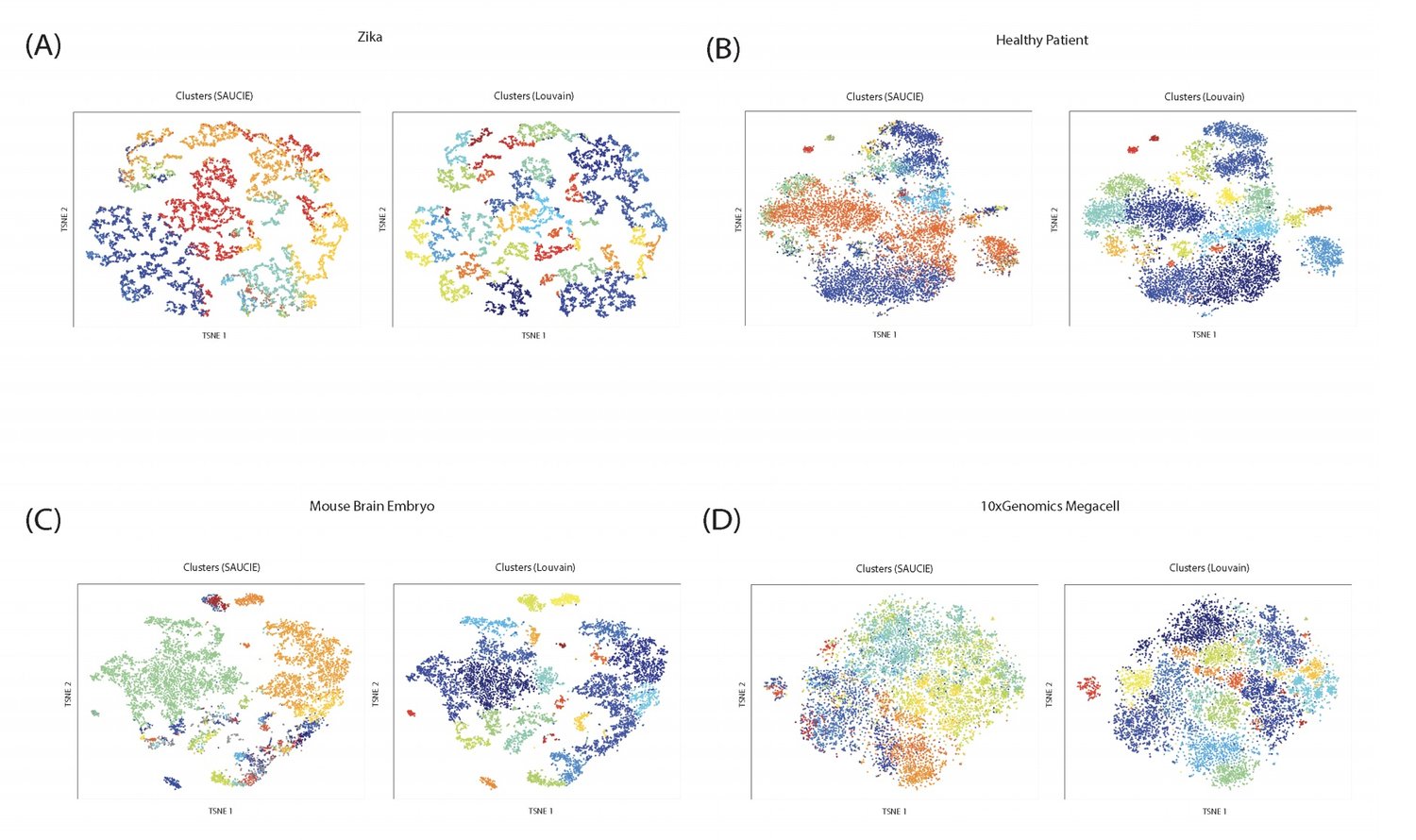
Handling the vast amounts of single-cell RNA-sequencing and CyTOF data, which are now being generated in patient cohorts, presents a computational challenge due to the noise, complexity, sparsity and batch effects present. Here, we propose a unified deep neural network-based approach to automatically process and extract structure from these massive datasets.
Our unsupervised architecture, called SAUCIE (Sparse Autoencoder for Unsupervised Clustering, Imputation, and Embedding), simultaneously performs several key tasks for single-cell data analysis including 1) clustering, 2) batch correction, 3) visualization, and 4) denoising/imputation. SAUCIE is trained to recreate its own input after reducing its dimensionality in a 2-D embedding layer which can be used to visualize the data.
Additionally, SAUCIE uses two novel regularizations: (1) an information dimension regularization to penalize entropy as computed on normalized activation values of the layer, and thereby encourage binary-like encodings that are amenable to clustering and (2) a Maximal Mean Discrepancy penalty to correct batch effects. Thus SAUCIE has a single architecture that denoises, batch-corrects, visualizes and clusters data using a unified representation. We show results on artificial data where ground truth is known, as well as mass cytometry data from dengue patients, and single-cell RNA-sequencing data from embryonic mouse brain.